Research Overview
Research Approaches
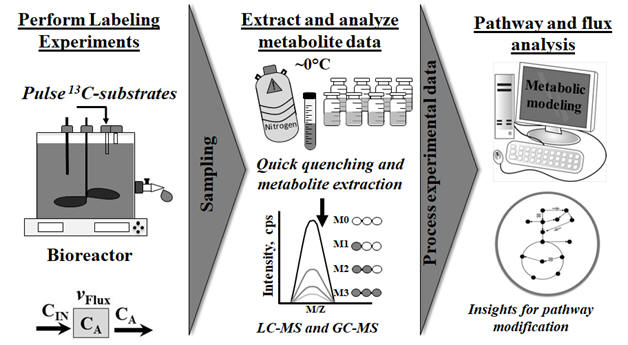
Metabolic analysis, process modeling and engineering: The Tang lab applies process or metabolic modeling, omics analysis, and 13C-Metabolic Flux Analysis (MFA) to decipher microbial metabolism, perform genome-to-phenome mapping, and optimize bioprocesses. The metabolic models integrated with omics data can also reveal complex cellular regulations. Our Lab projects focus on: 1) novel pathways in nonmodel microbial species; 2) flux network for using or co-utilizing different substrates; 3) flux organizations in engineered or evolved strains under stressed conditions, and 4) simulation or prediction of complex cellular processes. Recently, the Tang lab is working on knowledge mining and AI approaches to guide biomanufacturing development.

1) Multi-level and interconnected metabolic regulations: High throughput engineering tools and omics analyses have been well-established. However, it is still difficult to decipher complex cellular performance under various growth conditions. How to integrate metabolic models with data driven approaches to improve our understandings and designs of microbial metabolism for bioproductions?
2) Host genetic/nongenetic
heterogeneities: How to understand metabolic variations in single
cells? How
engineered strains are evolved in bioreactor conditions? Where are mutational locations? How to correlate
the growth stresses and metabolic burdens to
mutational driving force?
How to design strains to be
tolerant of metabolic burden?
3) Metabolite channeling: native strain may coordinate cascade enzymes via metabolite channels or metabolons. Such regulatory strategies reduce intermediate toxicity and increase enzyme efficiency, which lead to robust cellular functions under stressed conditions. It is still unclear how cells control degree of channeling in its central pathways and how channeling governs flux network.
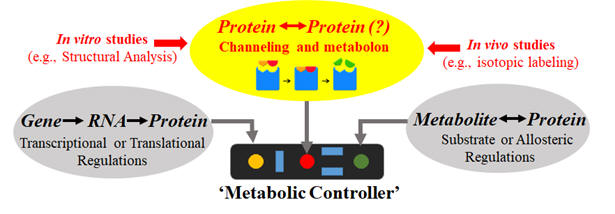
Fig 2: Cell metabolic regulations (Channeling is often overlooked.)
O
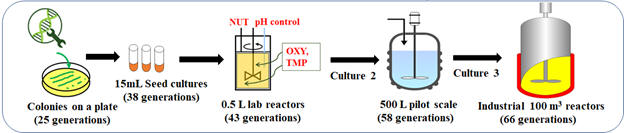
Fig. 3: Scale up of engineered hosts (over generations, non-producers will become dominant.)
__________________________________________________________________
Project 2. Study on proximity channeling: Metabolic models seldom consider intracellular diffusion limitations. Intriguingly, cascade enzymes of long-step pathways might self-assembled to form proximity channeling via weak interactions. However, proximity channeling of central pathways in living cells is difficult to verify and its roles on metabolic regulations are unclear. We are interested in deciphering proximity channeling and interactions between cascade enzymes in central pathways via combined 13C-metabolomics, flux modeling, metabolomics, in vitro enzyme assay, and targeted genetic engineering. See our recent papers (AICHE Journal 2019; Metabolic Engineering 2019; Biotech Advance 2018).
Project 3. Pathway
discovery and metabolic analysis of nonmodel species or genetic engineered
workhorses:
We have
characterized over 20 microbial species, including cyanobacteria,
Shewanella, Mycobacteria, Geobacter, Geobacillus, E.coli, Yarrow,
Sphingobium, Dehalococcoides, Thermoanaerobacter,
Heliobacteria, Roseobacter, Chlorobaculum, Chloroflexus, Chlorella, Clostridium,
Pseudomonas
Project 4. Machine learning for biomanufacturing: We work on knowledge mining and machine learning of metabolic engineering and bioprocess data. We aim to extract new insights from large datasets using natrual language processing (GPT-4). We are developing machine learning methods to improve model predictions and to optimize bioprocess controls. Please see papers on PlosOne (vol 14, e0210558), Biotech Advance (vol 36, 1308-1315), and PlosComputational Biology (vol 12, e1004838, machine learning software mFlux).
Project 5. Environmental Bioremediation, Plastic/Lignin Waste
Upcycling and CO2 capture:
Past Industrial Collaborations
(2010-2011) "CO2 utilization via algal process." (Consortium for Clean Coal Utilization, Peabody Energy, Arch Coal, Ameren)
(2011-2012) "Integration of anaerobic digestion with free fatty acid production via engineered microbial species" (Gates Foundation)
(2012-2013) "Microcoleus vaginatus cultivation for bio-fertilizers" (Terra Biologics)
(2014~2015) "Lignin Degradation using a soil bacterium" (Sandia National Lab)
(2015~2016) "Biofuel production from agricultural wastes and biogas" (Helee, LLC)
(2016-2019) "Fermentation optimization of yeast strains to produce natural nutrition" (Arch Innotek through NSF and NIH STTR grants)
(2021~2022) "Modeling and analysis of silk protein bio-productions" (RPI/Convergence Accelerator)